SBML4Humans GSoC Project Preparation (Week 1 Progress Update)
The Systems Biology Markup Language (SBML) is a standard language to represent biological models in a computer readable and exchangeable format. The goal of this project is to create a human-readable, reactive and navigable web-based interface for easy visualization of these SBML models. More information about the project can be found on the Github issue (https://github.com/nrnb/GoogleSummerOfCode/issues/164). I am preparing myself for this project and am being mentored by Dr. Matthias König.
The first week of learning taught me some great things, some new and some known (e.g. the SBML 3 structure, history, abstract and core ideas). I have gone through the following resources have reached a good understanding of the core ideas:
The SBML Homepage (http://sbml.org/Main_Page)
The SBML 3 abstract and overview in the following publication https://www.embopress.org/doi/full/10.15252/msb.20199110
Keating SM, Waltemath D, König M, Zhang F, Dräger A, Chaouiya C, Bergmann FT, Finney A, Gillespie CS, Helikar T, Hoops S, Malik-Sheriff RS, Moodie SL, Moraru II, Myers CJ, Naldi A, Olivier BG, Sahle S, Schaff JC, Smith LP, Swat MJ, Thieffry D, Watanabe L, Wilkinson DJ, Blinov ML, Begley K, Faeder JR, Gómez HF, Hamm TM, Inagaki Y, Liebermeister W, Lister AL, Lucio D, Mjolsness E, Proctor CJ, Raman K, Rodriguez N, Shaffer CA, Shapiro BE, Stelling J, Swainston N, Tanimura N, Wagner J, Meier-Schellersheim M, Sauro HM, Palsson B, Bolouri H, Kitano H, Funahashi A, Hermjakob H, Doyle JC, Hucka M; SBML Level 3 Community members. SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol. 2020 Aug;16(8):e9110. doi: 10.15252/msb.20199110. PMID: 32845085.The SBML Level 3 Version 2 specification in the document http://sbml.org/Special/specifications/sbml-level-3/version-2/core/release-2/sbml-level-3-version-2-release-2-core.pdf
Started going through the code base in the sbmlutils GitHub repository (https://github.com/matthiaskoenig/sbmlutils) and documentation (https://sbmlutils.readthedocs.io/en/latest/)
Studying the resources provided a good overview of the core ideas of SBML and I got a good overview of its needs, use cases, its basic functionality, hierarchy of different constructs and various object-oriented features.
In summary, SBML is a language to convert real-world or hypothetical biological, biochemical or biophysical models into a computer understandable and shareable format. It is similar to HTML, but not for developing websites but for developing biological models. The encoding in XML provides a good idea of the tree-like structure and hierarchy that such biological systems hold in their essence. SBML gives a specification to represent information about the biological models. Tools can then be built around this specification to visualize the information, which would have been difficult to do on a computer. The aim of this project is also to develop one such tool to present the information in a human-understandable and navigable format.
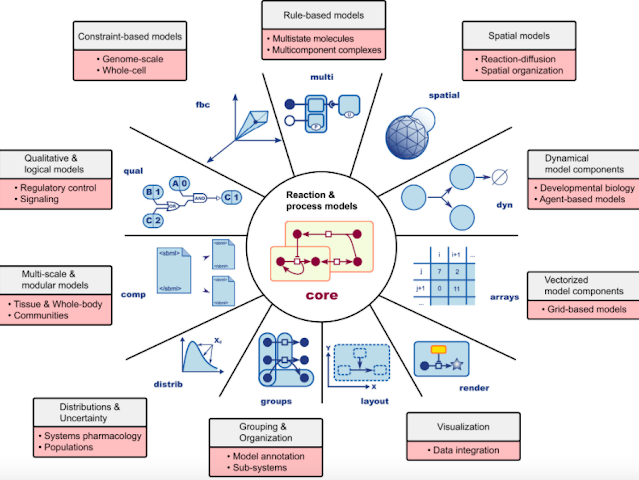
The core of SBML is built in such a way that it can be used differently for different use-cases but at the same time maintaining a uniformity across all such applications due to the common core. It encapsulates the object-oriented programming (OOP) principle of inheritance and abstraction. The core provides one with all the essential utilities to properly and completely model most biological systems.
We build our models based on the core objects. The core provides the required building blocks to represent real-world entities (eg. reactants, containers, reactions, kinetics, etc.) in a computer-understandable form. The relationships between these elements (eg. reactants are stored in the containers, reactants take part in the reactions, etc) are also specified using the core objects and their attributes. The core model objects are provided in the SBML core.
Specialization on top of these core models are provided by packages. The applications of these models can be decided by the researcher or developer. Someone may want to make a probabilistic or statistical study on the concentration of a particular species in a certain reaction : the distributions package can be used here. Similarly, to make a logical output analysis of the concentration of ions in the synaptic gap of two neurons, one can use the logical and qualitative analysis models.
The basic workflow for creating the outline of a model is to think about its participating species (reactants, products, catalysts), compartments, reactions, mathematical rules and rate laws, etc. These elements form the basic structure of most biological models represented using SBML. Such a core model can then be stored in the required format, analysed, shared and be further developed in the future.
I have got a good and comprehensive idea of the constructs, their syntaxes, related attributes, the meaning of those attributes, parent-child relationships, etc. I have a few doubts regarding some concepts in annotation, but I’ll keep on revisiting it and hope that it will get clear. The UML diagrams, their relationships, and flow of information were explained in a clear manner.
The in-section examples and seventh section in the specification document explained many examples which further strengthened my understanding of the above elements. I got a clear idea of entire document specifications, element declarations and definitions, function definitions just like the programs I have worked on before in languages like C, C++, Java or Python. One of the major learning takeaways from this exercise was to understand the use of the various OOP principles in the formulation of the documentation structure and meaning of the elements. The mapping of the models to their real-world realizations was cleared a lot by these examples and principles.
I have started studying the codebase on Github (https://github.com/matthiaskoenig/sbmlutils) and also the documentation (https://sbmlutils.readthedocs.io/en/latest/). I would now start testing it on my local system and get more familiarized with the code, development, testing and use cases. I would keep on visiting these first resources from time to time and gain more mastery over these topics which would help me in the future stages.




Comments
Post a Comment